What is a gene?
Any region of DNA that codes for a specific amino acid sequence-->forms RNA/polypeptides.
...
What is a fish?
beeeeeeep.
Tuesday, March 1, 2011
Mutations
A. Substitutions
1. Silent mutation
-no effect on overall structure
-e.g. UUU and UUC both code for Phe
2. Missense mutation
-code for a different type of amino acid
e.g. UUU-->UUG Phe-->Leu
3. Nonsense mutation
-amino acid to non-functioning protein/early stop
-e.g. UAC-->UAG Tyr-->Stop
Example:
Wildtype: AUG CUU UGC CCU UAA
M L C P stop
Missense: AUG CUU UGG CCU UAA
M L T P
Nonsense: AUG CUU UGA CCU UAA
M L stop
B. Frameshifts
4. Insertion
-addition of nucleotide(s)
5. Deletion
-loss of nucleotide(s)
Example:
Wildtype: THE FAT CAT ATE THE RAT
Insertion: THE FAR TCA TAT ETH ERA T
Deletion: THE FAC ATA TET HER AT
1. Silent mutation
-no effect on overall structure
-e.g. UUU and UUC both code for Phe
2. Missense mutation
-code for a different type of amino acid
e.g. UUU-->UUG Phe-->Leu
3. Nonsense mutation
-amino acid to non-functioning protein/early stop
-e.g. UAC-->UAG Tyr-->Stop
Example:
Wildtype: AUG CUU UGC CCU UAA
M L C P stop
Missense: AUG CUU UGG CCU UAA
M L T P
Nonsense: AUG CUU UGA CCU UAA
M L stop
B. Frameshifts
4. Insertion
-addition of nucleotide(s)
5. Deletion
-loss of nucleotide(s)
Example:
Wildtype: THE FAT CAT ATE THE RAT
Insertion: THE FAR TCA TAT ETH ERA T
Deletion: THE FAC ATA TET HER AT
Translation :S
Translation: Ribosomes use mRNA as blueprint to synthesize amino acids which form proteins.
-64 codon combinations
-61 code for amino acids
-3 code for stop codon
-20 amino acids
-Wobble Effect: 3rd position of codon is flexible
-->prevents mutations
1. Initiation
i. Ribosome recognizes the 5' cap and binds to it. It contains a large and a small subunit.
ii. Ribosome moves along mRNA.
iii. Translation begins when ribosome reads AUG (start codon).
iv. tRNA delivers Met to the P-site. There is an anti-codon on the tRNA, which is complementary to the codon on mRNA.
2. Elongation
i. A 2nd tRNA carrying an amino acid enters the A-site. Hydrogen bond is formed between the codon and the anicodon (by elongation factors).
ii. Peptide bond is formed between the amino acids.
iii. The petide chain is transfered to the A-site. 1st tRNA leaves the P-site.
iv. tRNA moves from A-site to P-site with the codon (translocation). New codon is available at the A site. 1st tRNA leaves the ribosome at the E-site.
Note: mRNA is read from 5' to 3', codon by codon.
3. Termination
i. Translation stops when the ribosome reads the stop codon (UGA, UAG, UAA).
ii. Release factor hydrolyzes the bond between the polypeptide and the tRNA at the P site.
iii. The polypetide is released :)
-64 codon combinations
-61 code for amino acids
-3 code for stop codon
-20 amino acids
-Wobble Effect: 3rd position of codon is flexible
-->prevents mutations
1. Initiation
i. Ribosome recognizes the 5' cap and binds to it. It contains a large and a small subunit.
ii. Ribosome moves along mRNA.
iii. Translation begins when ribosome reads AUG (start codon).
iv. tRNA delivers Met to the P-site. There is an anti-codon on the tRNA, which is complementary to the codon on mRNA.
2. Elongation
i. A 2nd tRNA carrying an amino acid enters the A-site. Hydrogen bond is formed between the codon and the anicodon (by elongation factors).
ii. Peptide bond is formed between the amino acids.
iii. The petide chain is transfered to the A-site. 1st tRNA leaves the P-site.
iv. tRNA moves from A-site to P-site with the codon (translocation). New codon is available at the A site. 1st tRNA leaves the ribosome at the E-site.
Note: mRNA is read from 5' to 3', codon by codon.
3. Termination
i. Translation stops when the ribosome reads the stop codon (UGA, UAG, UAA).
ii. Release factor hydrolyzes the bond between the polypeptide and the tRNA at the P site.
iii. The polypetide is released :)
Transcription :S
Transcription: Copying of genetic information from DNA to mRNA
1. Initiation:
TATA box, transcriptioin factors (TFs), and RNA polymerase II form the initiation complex upstream of the DNA molecule.
Upstream --> Promoter (sequence of DNA)
i. TATA box is present and is recognized by transcription factors (proteins) in eukaryotes.
ii. RNA polymerase binds to the transcription factors.
iii. Initiation complex is formed.
iv. Initiation complex signals transcription.
2. Elongation
i. RNA polymerase binds to the promoter and unwinds the DNA double helix.
ii. DNA template is read from 3' to 5', and pre-mRNA strand elongates from 5' to 3'.

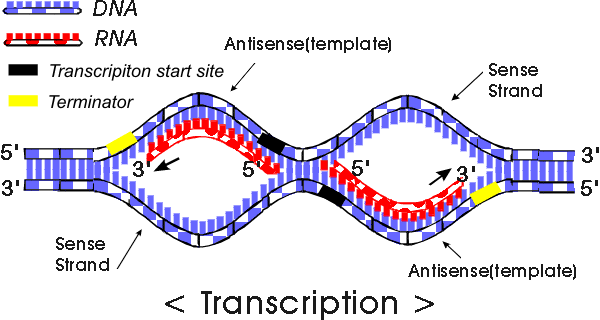
coding strand=sense=non-template
-not used for transcription
-similar to pre-mRNA, except pre-mRNA has U instead of T
non-coding strand=antisense=template
-complementary to pre-mRNA
-read from 3' to 5'
Note: TATA box is not transcribed.
3. Termination
i. Transcription stops at AAUAAA
4. Modifications
i. 5' cap
-protects mRNA from hydrolytic enzymes
-"attach here" signal for ribosomes
ii. poly-A tail at the 3' end
-inhibite hydrolysis
-facilitate ribosome attachment
-facilitate export of mRNA
iii. RNA splicing
introns=non-coding regions
extrons=code for amino acids
-pre-mRNA+snRNP+proteins-->spliceosomes
-snRNA acts as rybozyme and pairs with nucleotides at the ends of introns
-introns form loops and are excised
-exons are spliced together

1. Initiation:
TATA box, transcriptioin factors (TFs), and RNA polymerase II form the initiation complex upstream of the DNA molecule.
Upstream --> Promoter (sequence of DNA)
i. TATA box is present and is recognized by transcription factors (proteins) in eukaryotes.
ii. RNA polymerase binds to the transcription factors.
iii. Initiation complex is formed.
iv. Initiation complex signals transcription.
2. Elongation
i. RNA polymerase binds to the promoter and unwinds the DNA double helix.
ii. DNA template is read from 3' to 5', and pre-mRNA strand elongates from 5' to 3'.


coding strand=sense=non-template
-not used for transcription
-similar to pre-mRNA, except pre-mRNA has U instead of T
non-coding strand=antisense=template
-complementary to pre-mRNA
-read from 3' to 5'
Note: TATA box is not transcribed.
3. Termination
i. Transcription stops at AAUAAA
4. Modifications
i. 5' cap
-protects mRNA from hydrolytic enzymes
-"attach here" signal for ribosomes
ii. poly-A tail at the 3' end
-inhibite hydrolysis
-facilitate ribosome attachment
-facilitate export of mRNA
iii. RNA splicing
introns=non-coding regions
extrons=code for amino acids
-pre-mRNA+snRNP+proteins-->spliceosomes
-snRNA acts as rybozyme and pairs with nucleotides at the ends of introns
-introns form loops and are excised
-exons are spliced together

Saturday, February 19, 2011
10 things about replication
Overview:
1. The replication process is semiconservative.
2. Hydrogen bonds between complementary bases break.
3. DNA helix unzip. Replication bubbles are formed.

4. Each single-stranded DNA sequence acts as a guiding pattern for producing a complementary DNA strand (template).
5. Because the 2 strands in a double helix are antiparallel, one goes in the 5' to 3' direction while the other goes in the 3' to 5' direction.
6. However, DNA only elongates in the 5' to 3' direction.
7. The leading strand elongates in the 5' to 3' direction towards the replication fork.
8. Okazaki fragments are formed on the lagging strand. They elongate in the 5' to 3' direction, away from the replication fork.
9. THE TOP STRAND IS NOT NECESSARILY THE LEADING STRAND.
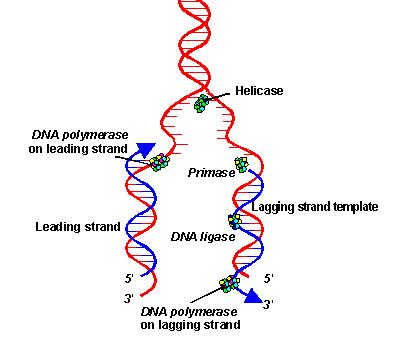
10. The stages are initiation, elongation, termination, and repair.
The stages:
i. Initiation - separating the DNA strands
Helicase unwinds the double-stranded DNA by breaking the hydrogen bonds.
Replication bubbles are formed.
Single-stranded binding proteins stabilize the single-stranded DNA.
Gyrase release tension brought by unwinding of DNA strands by cutting and resealing the strands.
RNA primase produce primers, which signal the release of DNA polymerase III.
RNA primers are recognized by DNA polymerase III. They anneal to the DNA template.
ii. Elongation - Building the complementary strands
RNA primers act as starting points of replication.
DNA polymerase III recognize the RNA primers. They add nucleotides to the template.
Nucleotides are continuously being added to the 3' end of the elongating strand. The strand elongates in the 5' to 3' direction.
The leading strand elongates towards the replication fork.
Okazaki fragments are formed on the lagging strand.

iii. Termination - Joining the DNA
DNA polymerase I replace RNA primers with DNA.
DNA ligase join the gaps in DNA.
Okazaki fragments are bonded by phosphodiester bonds.
iv. Repair - Proof-reading the DNA
DNA polymerase I and III act as exonuclease which proof-read the complenentary strand.
The strands automatically twist into a double helix.
1. The replication process is semiconservative.
2. Hydrogen bonds between complementary bases break.
3. DNA helix unzip. Replication bubbles are formed.

4. Each single-stranded DNA sequence acts as a guiding pattern for producing a complementary DNA strand (template).
5. Because the 2 strands in a double helix are antiparallel, one goes in the 5' to 3' direction while the other goes in the 3' to 5' direction.
6. However, DNA only elongates in the 5' to 3' direction.
7. The leading strand elongates in the 5' to 3' direction towards the replication fork.
8. Okazaki fragments are formed on the lagging strand. They elongate in the 5' to 3' direction, away from the replication fork.
9. THE TOP STRAND IS NOT NECESSARILY THE LEADING STRAND.

10. The stages are initiation, elongation, termination, and repair.
The stages:
i. Initiation - separating the DNA strands
Helicase unwinds the double-stranded DNA by breaking the hydrogen bonds.
Replication bubbles are formed.
Single-stranded binding proteins stabilize the single-stranded DNA.
Gyrase release tension brought by unwinding of DNA strands by cutting and resealing the strands.
RNA primase produce primers, which signal the release of DNA polymerase III.
RNA primers are recognized by DNA polymerase III. They anneal to the DNA template.
ii. Elongation - Building the complementary strands
RNA primers act as starting points of replication.
DNA polymerase III recognize the RNA primers. They add nucleotides to the template.
Nucleotides are continuously being added to the 3' end of the elongating strand. The strand elongates in the 5' to 3' direction.
The leading strand elongates towards the replication fork.
Okazaki fragments are formed on the lagging strand.

iii. Termination - Joining the DNA
DNA polymerase I replace RNA primers with DNA.
DNA ligase join the gaps in DNA.
Okazaki fragments are bonded by phosphodiester bonds.
iv. Repair - Proof-reading the DNA
DNA polymerase I and III act as exonuclease which proof-read the complenentary strand.
The strands automatically twist into a double helix.
Friday, February 11, 2011
Vocabs.
1. Cytosol
A semi-liquid found in the cytoplasm that contains microfillaments of the cytoskeleton (the supporting structure). It is where most cellular metabolism occurs.
2. Germ cells
Reproductive cells.
3. Diploid
A cell that contains 2 pairs of chromosomes (2n). For humans (under normal conditions), n=23, so 2n=46.
4. Hapoid
A cell that contains 1 complete set of chromosomes (n). For humans (under normal conditions), n=23.
5. Cytokinesis
A process in mitosis. It takes place in telophase. A cleavage furrow is formed at the center of the cell. The cell is then divided into 2 identical cells.
http://highered.mcgraw-hill.com/olcweb/cgi/pluginpop.cgi?it=swf::535::535::/sites/dl/free/0072437316/120073/bio14.swf::Mitosis%20and%20Cytokinesis
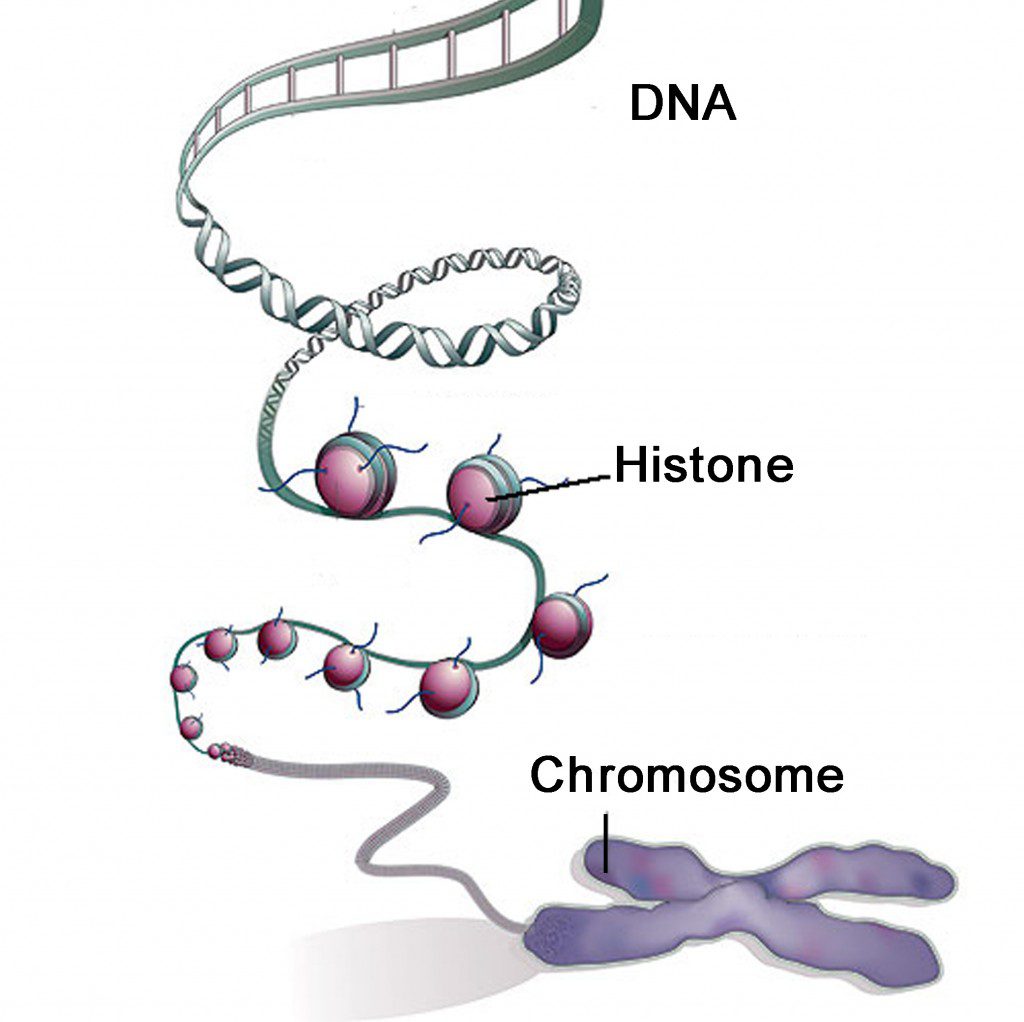
6. Histones
Proteins that are responsible for the structure of the chromotin and packaging of DNA into chromosomes.
7. Cell plates
A structure that is developed at the center of the cell during cytokinesis. It separates the chromosomes.

8. Chromatin
A mass that contains DNA and protein that will condense to form chromosomes during mitosis.
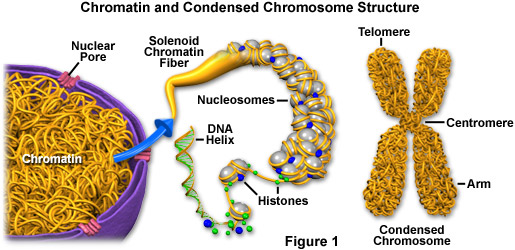
9. Sister chromatids
Single-stranded chromosomes that are identical and are connected by a centromere.

10. Homologs
One of a pair of chromosomes. In a pair of chromosomes, one homolog is from father and the other is from mother.
11. mRNA
Responsible for protein synthesis. It is attached to the DNA template during transcription. During translation, it binds to ribosome (sugar). At the same time, it serves as the binding site for tRNA. tRNA attaches 3 nucleotides to the corresponding sites on the mRNA.
http://www.youtube.com/watch?v=NJxobgkPEAo&feature=related
12. Prokaryotes
-ancient
-do not have cell nuclei
-do not have organelles surrounded by membranes
-DNA is concentrated in the nucleoid, but no membrane separates this part from the rest of the cell
13. Eukaryotes
-contains a cell nuclei
-have organelles surrounded by membranes
-some contains chloroplasts
-DNA is concentrated in the nucleus
A semi-liquid found in the cytoplasm that contains microfillaments of the cytoskeleton (the supporting structure). It is where most cellular metabolism occurs.

2. Germ cells
Reproductive cells.
3. Diploid
A cell that contains 2 pairs of chromosomes (2n). For humans (under normal conditions), n=23, so 2n=46.
4. Hapoid
A cell that contains 1 complete set of chromosomes (n). For humans (under normal conditions), n=23.
5. Cytokinesis
A process in mitosis. It takes place in telophase. A cleavage furrow is formed at the center of the cell. The cell is then divided into 2 identical cells.
http://highered.mcgraw-hill.com/olcweb/cgi/pluginpop.cgi?it=swf::535::535::/sites/dl/free/0072437316/120073/bio14.swf::Mitosis%20and%20Cytokinesis
6. Histones
Proteins that are responsible for the structure of the chromotin and packaging of DNA into chromosomes.

7. Cell plates
A structure that is developed at the center of the cell during cytokinesis. It separates the chromosomes.

8. Chromatin
A mass that contains DNA and protein that will condense to form chromosomes during mitosis.

9. Sister chromatids
Single-stranded chromosomes that are identical and are connected by a centromere.

10. Homologs
One of a pair of chromosomes. In a pair of chromosomes, one homolog is from father and the other is from mother.
11. mRNA
Responsible for protein synthesis. It is attached to the DNA template during transcription. During translation, it binds to ribosome (sugar). At the same time, it serves as the binding site for tRNA. tRNA attaches 3 nucleotides to the corresponding sites on the mRNA.
http://www.youtube.com/watch?v=NJxobgkPEAo&feature=related
12. Prokaryotes
-ancient
-do not have cell nuclei
-do not have organelles surrounded by membranes
-DNA is concentrated in the nucleoid, but no membrane separates this part from the rest of the cell
13. Eukaryotes
-contains a cell nuclei
-have organelles surrounded by membranes
-some contains chloroplasts
-DNA is concentrated in the nucleus
Tuesday, February 8, 2011
Geneticists
1. T. H. Morgan
-observed eye colors of fruit flies
Reference: http://www.nature.com/scitable/topicpage/thomas-hunt-morgan-and-sex-linkage-452
-all white-eyed flies were males
-the white-eye allele is recessive and is located on the X chromosome
-Males have white eyes because they only have 1 X chromosome. If the white-eye allele is inherited, it becomes dominant. (Well, not exactly dominant. But that's the only allele for eye color it has, right?)
Theories:
i. Genes are located on chromosomes
ii. Either protein or DNA codes for the genetic information
2. Hershey and Chase
The experiment:
Purpose: To illustrate the relation between DNA and heredity and to demonstrate that DNA is the genetic material
Procedure:
i. Allow phages to attack a bacterial cell. Their proteins are radioactively labelled.
ii. Allow a second group of phages to infect a bacterial cell. Their DNA is radioactively labelled.
Result:
i. Phages produced by the 1st cell were not radioactive.
ii. Phages produced by the 2nd cell were radioactive
Conclusion:
DNA is the genetic material that is passed from generation to generation.
check this out!!
http://highered.mcgraw-hill.com/olc/dl/120076/bio21.swf
3. Frederick Griffith
The experiment of transformation:
R strain --> rough --> harmless
S strain --> smooth --> fetal
Procedure:
Inject the strain(s) into mice
Trial 1: S strain is injected
Trial 2: R strain is injected
Trial 3: S strain is killed (by heat) and injected
Trial 4: Dead S strain and alive R strain are injected
Hypothesis:
Trial 1: The mouse would die.
Trial 2: The mouse would live.
Trial 3: The mouse would live.
*Trial 4: The mouse would live.
Results:
Trial 1: The mouse died.
Trial 2: The mouse lived.
Trial 3: The mouse lived.
*Trial 4: The mouse died.
Conclusion:
The injection caused a change in genotype and phenotype. This process is called transformation.

4. Erwin Chargaff
-discovered the 4 bases of DNA
Purines (double-ringed): adenine, guanine
Pyrimidines (single-ringed): thymine, cytosine
% T = % A
% G = % C
Each species has a distinct DNA composition.

5. Maurice Wilkins and Rosalind Franklin
-studied the structure of DNA according the diffraction pattern of X-ray
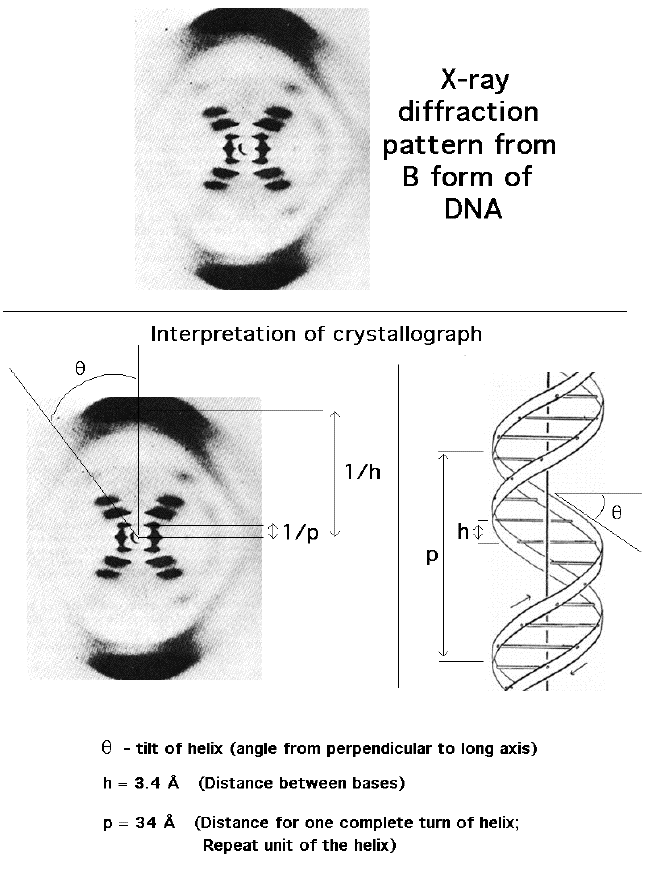
6. Watson and Crick

-studied Wilkins and Franklin's research
-DOUBLE HELIX
-Purine + Pyrimidine
-A=T
-G≡C
-diameter = 2 nm

-observed eye colors of fruit flies
| Cross | Outcome | |
| Expected Phenotypes | Observed Phenotypes | |
| P1 Red ♀ × P1 White ♂ | F1 = All Red | F1 = All Red* |
| F1 Red ♀ × F1 Red ♂ | 75% Red ♀ and ♂ 25% White ♀ and ♂ | 50% Red ♀ 25% Red ♂ 25% White ♂ |
Reference: http://www.nature.com/scitable/topicpage/thomas-hunt-morgan-and-sex-linkage-452
-all white-eyed flies were males
-the white-eye allele is recessive and is located on the X chromosome
-Males have white eyes because they only have 1 X chromosome. If the white-eye allele is inherited, it becomes dominant. (Well, not exactly dominant. But that's the only allele for eye color it has, right?)
Theories:
i. Genes are located on chromosomes
ii. Either protein or DNA codes for the genetic information
2. Hershey and Chase
The experiment:
Purpose: To illustrate the relation between DNA and heredity and to demonstrate that DNA is the genetic material
Procedure:
i. Allow phages to attack a bacterial cell. Their proteins are radioactively labelled.
ii. Allow a second group of phages to infect a bacterial cell. Their DNA is radioactively labelled.
Result:
i. Phages produced by the 1st cell were not radioactive.
ii. Phages produced by the 2nd cell were radioactive
Conclusion:
DNA is the genetic material that is passed from generation to generation.
check this out!!
http://highered.mcgraw-hill.com/olc/dl/120076/bio21.swf
3. Frederick Griffith
The experiment of transformation:
R strain --> rough --> harmless
S strain --> smooth --> fetal
Procedure:
Inject the strain(s) into mice
Trial 1: S strain is injected
Trial 2: R strain is injected
Trial 3: S strain is killed (by heat) and injected
Trial 4: Dead S strain and alive R strain are injected
Hypothesis:
Trial 1: The mouse would die.
Trial 2: The mouse would live.
Trial 3: The mouse would live.
*Trial 4: The mouse would live.
Results:
Trial 1: The mouse died.
Trial 2: The mouse lived.
Trial 3: The mouse lived.
*Trial 4: The mouse died.
Conclusion:
The injection caused a change in genotype and phenotype. This process is called transformation.

4. Erwin Chargaff
-discovered the 4 bases of DNA
Purines (double-ringed): adenine, guanine
Pyrimidines (single-ringed): thymine, cytosine
% T = % A
% G = % C
Each species has a distinct DNA composition.

5. Maurice Wilkins and Rosalind Franklin
-studied the structure of DNA according the diffraction pattern of X-ray

6. Watson and Crick

-studied Wilkins and Franklin's research
-DOUBLE HELIX
-Purine + Pyrimidine
-A=T
-G≡C
-diameter = 2 nm
Subscribe to:
Posts (Atom)