Transcription: Copying of genetic information from DNA to mRNA
1. Initiation:
TATA box,
transcriptioin factors (TFs), and
RNA polymerase II form the initiation complex upstream of the DNA molecule.
Upstream --> Promoter (sequence of DNA)
i.
TATA box is present and is recognized by transcription factors (proteins) in eukaryotes.
ii.
RNA polymerase binds to the transcription factors.
iii.
Initiation complex is formed.
iv. Initiation complex signals transcription.
2. Elongation
i.
RNA polymerase binds to the promoter and unwinds the DNA double helix.
ii. DNA template is
read from 3' to 5', and pre-mRNA strand
elongates from 5' to 3'.

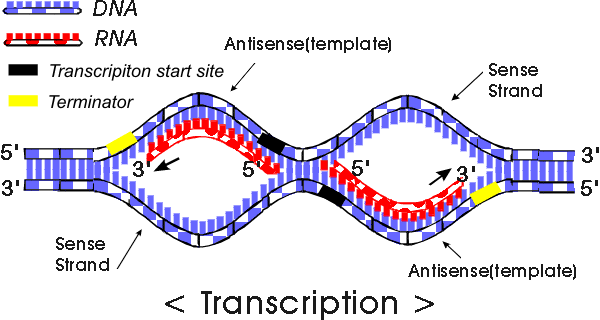
coding strand=sense=non-template
-not used for transcription
-similar to pre-mRNA, except pre-mRNA has U instead of T
non-coding strand=antisense=template
-complementary to pre-mRNA
-read from 3' to 5'
Note: TATA box is not transcribed.
3. Termination
i. Transcription stops at
AAUAAA
4. Modifications
i.
5' cap
-protects mRNA from hydrolytic enzymes
-"attach here" signal for ribosomes
ii.
poly-A tail at the 3' end
-inhibite hydrolysis
-facilitate ribosome attachment
-facilitate export of mRNA
iii.
RNA splicing
introns=non-coding regions
extrons=code for amino acids
-pre-mRNA+snRNP+proteins-->spliceosomes
-snRNA acts as rybozyme and pairs with nucleotides at the ends of introns
-introns form loops and are excised
-exons are spliced together



